Classifiers, table of contents
This section describes perClass classifiers based on assumption of Gaussian distribution.
- 13.2.1. Introduction
- 13.2.2. Nearest mean classifier
- 13.2.2.1. Scaled nearest mean
- 13.2.3. Linear discriminant assuming normal densities
- 13.2.4. Quadratic classifier assuming normal densities
- 13.2.5. Gaussian model or classifier
- 13.2.6. Constructing Gaussian model from parameters
- 13.2.7. Generating data based on Gaussian model
- 13.2.8. Gaussian mixture models
- 13.2.8.1. Automatic estimation of number of mixture components
- 13.2.8.2. Choosing number of mixture components manually
- 13.2.8.3. Clustering data using a mixture model
- 13.2.9. Regularization of Gaussian models
13.2.1. Introduction ↩
The family of Gaussian classifiers assumes that the observations are generated by a random process that has normal distribution. Density function of a normal distribution is defined by mean vector, and covariance matrix. If multiple Gaussian models are used, they are additionally weighted by priors.
In the simplest situation, the assumption is that each class is defined by a single Gaussian density. This yields nearest mean, linear and quadratic classifier. More complex setup assumes that each class may be modeled by a mixture of several Gaussians. This results in Gaussian mixture models.
In general, classifiers based on assumption of normality estimate probability densities. Therefore, their use is in practice limited to lower-dimensional situations. If the number of samples is too low compared to dimensionality or data exhibit strong subspace structure, normal models have difficulties in inverting covariance matrices.
13.2.2. Nearest mean classifier ↩
The nearest mean classifier sdnmean leverages assumption of normality.
It uses normal model with identity covariance matrix for all classes.
Nearest mean is one of the most simple classifiers useful in situations with few samples and large number of features.
>> load fruit
260 by 2 sddata, 3 classes: 'apple'(100) 'banana'(100) 'stone'(60)
>> p=sdnmean(a)
sequential pipeline 2x1 'Nearest mean+Decision'
1 Nearest mean 2x3 unit cov.mat.
2 Decision 3x1 weighting, 3 classes
>> sdscatter(a,p)
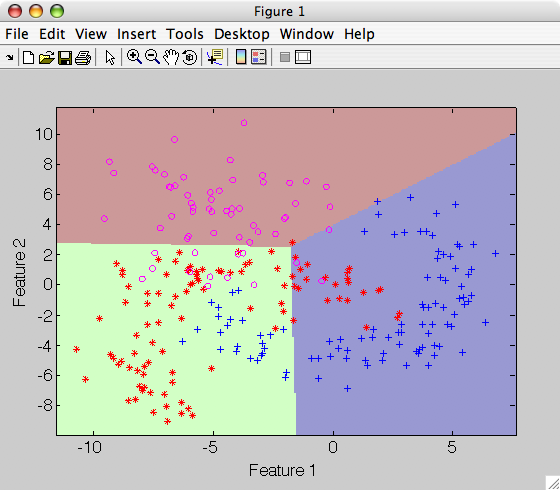
An alternative formulation of nearest mean using distances to prototypes is described here.
As for all Gaussian models, user may provide apriori-known class prior
probabilities when training sdnmean with 'priors' option. If not
provided, priors are estimated from training data set.
13.2.2.1. Scaled nearest mean ↩
With the 'scaled' option, sdnmean uses covariance matrices with a
separately estimated variance for each feature.
In this way, nearest mean classifier takes into account data scaling and will not change if the multiplicative scaling of features is applied.
>> p2=sdnmean(tr,'scaled')
sequential pipeline 2x1 'Nearest mean+Decision'
1 Nearest mean 2x3 scaled diag.cov.mat
2 Decision 3x1 weighting, 3 classes
13.2.3. Linear discriminant assuming normal densities ↩
sdlinear is a linear discriminant based on assumption of normal
densities. The pooled covariance matrix is computed by averaging per-class
covariances taking into account class priors.
>> load fruit;
>> a=a(:,:,[1 2])
260 by 2 sddata, 2 classes: 'apple'(100) 'banana'(100)
>> p=sdlinear(a)
sequential pipeline 2x1 'Gaussian model+Normalization+Decision'
1 Gaussian model 2x2 single cov.mat.
2 Normalization 2x2
3 Decision 2x1 weighting, 2 classes
Note the normalization step in the pipeline p. This assures that the
sdlinear soft output is posterior probability (confidence). Due to this
normalization, sdlinear requires two or more classes be present in the
input data set.
Confusion matrix estimated on the training set at default operating point:
>> sdconfmat(a.lab,a*p,'norm')
ans =
True | Decisions
Labels | apple banana | Totals
-------------------------------------
apple | 0.890 0.110 | 1.00
banana | 0.150 0.850 | 1.00
-------------------------------------
In order to use specific priors, provide them using the priors option:
>> p=sdlinear(a,'priors',[0.8 0.2])
sequential pipeline 2x1 'Gaussian model+Normalization+Decision'
1 Gaussian model 2x2 single cov.mat.
2 Normalization 2x2
3 Decision 2x1 weighting, 2 classes
>> sdconfmat(a.lab,a*p,'norm')
ans =
True | Decisions
Labels | apple banana | Totals
-------------------------------------
apple | 0.950 0.050 | 1.00
banana | 0.200 0.800 | 1.00
-------------------------------------
Note that by increasing the apple class prior we lower the apple error. However, the banana error will increase accordingly.
The sdlinear classifier may be regularized by adding a small constant to
a covariance diagonal. See the following section for an example on how to
use automatic regularization.
13.2.4. Quadratic classifier assuming normal densities ↩
sdquadratic implements quadratic discriminant based on assumption of
normal densities. In fact, sdquadrartic is composed of an sdgauss
pipeline with an extra normalization step that makes sure the classifier
returns posteriors. Due to this class normalization, sdquadratic can only
be trained on data set with two or more classes.
The quadratic decision boundary is achieved by estimating a specific covariance matrix for each class.
>> p=sdquadratic(a)
sequential pipeline 2x1 'Quadratic discriminant'
1 Gaussian model 2x2 full cov.mat.
2 Normalization 2x2
3 Decision 2x1 weighting, 2 classes
Detailed display of pipeline contents shows the outputs of the first and second step (density and posterior):
>> p'
sequential pipeline 2x1 'Quadratic discriminant'
1 Gaussian model 2x3 full cov.mat.
inlab: 'Feature 1','Feature 2'
lab: 'apple','banana','stone'
output: probability density
complab: component labels
2 Normalization 3x3
inlab: 'apple','banana','stone'
lab: 'apple','banana','stone'
output: posterior
3 Decision 3x1 weighting, 3 classes
inlab: 'apple','banana','stone'
output: decision ('apple','banana','stone')
Visualizing the classifier decisions:
>> sdscatter(a,p)
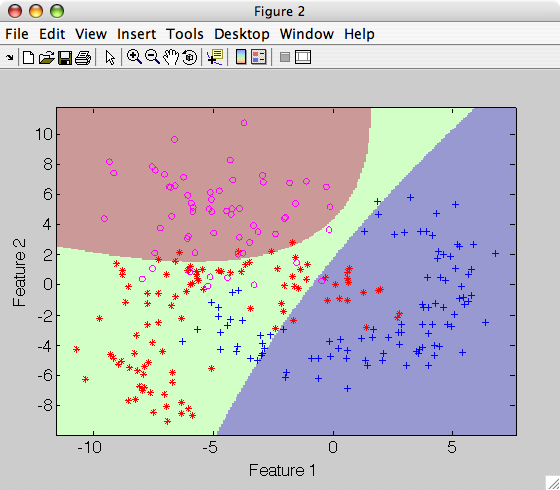
Similarly to other normal-based models, the user may fix class priors with 'priors' option, if know apriori.
13.2.5. Gaussian model or classifier ↩
sdgauss implements a general Gaussian model or classifier with full
covariances. By default, sdgauss returns a classifier:
>> p=sdgauss(a)
sequential pipeline 2x1 'Gaussian model+Decision'
1 Gaussian model 2x3 full cov.mat.
2 Decision 3x1 weighting, 3 classes
Unlike sdquadratic, sdgauss does not normalize class outputs. It
returns probability density and, therefore, may be used to build a detector.
In fact, training sdgauss on data with only one class returns a one-class
classifier that accepts all training examples:
>> a
'Fruit set' 260 by 2 sddata, 3 classes: 'apple'(100) 'banana'(100) 'stone'(60)
>> p=sdgauss(a(:,:,'apple'))
sequential pipeline 2x1 'Gaussian model+Decision'
1 Gaussian model 2x1 full cov.mat.
2 Decision 1x1 threshold on 'apple'
>> sdscatter(a,p)
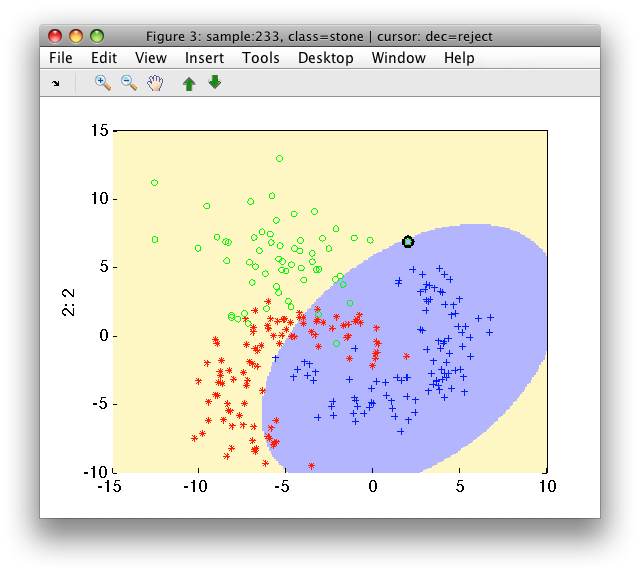
13.2.6. Constructing Gaussian model from parameters ↩
sdgauss may be also used to create Gaussian pipeline from parameters. We
need to provide:
sddataset with component means- cell array with covariance matrices
- vector of priors with an entry for each component
Example: I'd like to create a model with two components, one with mean at
[0 0] and the other at [5 3] with the first covariance unit and second
[1 0.3; -0.8 0.5].
>> m=sddata([0 0; 5 3])
2 by 2 sddata, class: 'unknown'
>> cov={eye(2) [1 .5; .5 2]}
cov =
[2x2 double] [2x2 double]
>> prior=[0.5 0.5]
prior =
0.5000 0.5000
>> p=sdgauss(m,cov,prior)
Gaussian model pipeline 2x1
To visualize the output of the pipline p, we need a data set to pass to
sdscatter. We can use class means and add two points extending our view:
>> sdscatter(sddata([+m; -5 -5; 10 10]),p)
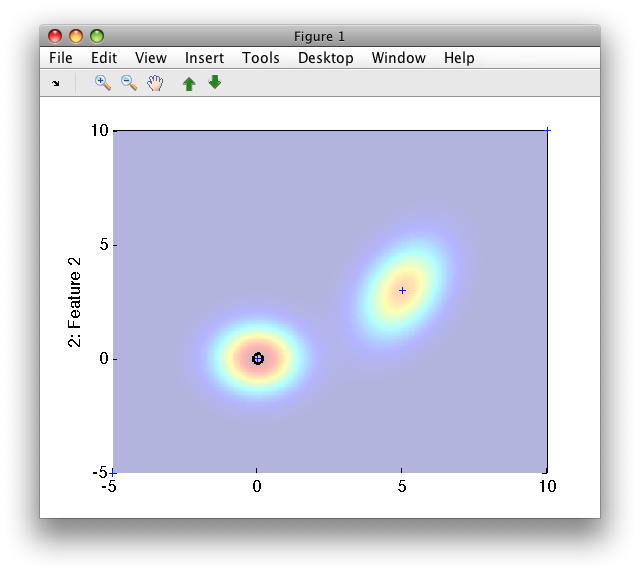
13.2.7. Generating data based on Gaussian model ↩
Gaussian model are "generative" which means we can use them to create a data set following the same distribution.
In perClass, we may use the sdgenerate command to do that. Using the
pipeline p form the previous section, we can create data set with 100
samples per component by:
>> b=sdgenerate(p,100)
200 by 2 sddata, class: 'unknown'
>> sdscatter(b,p)
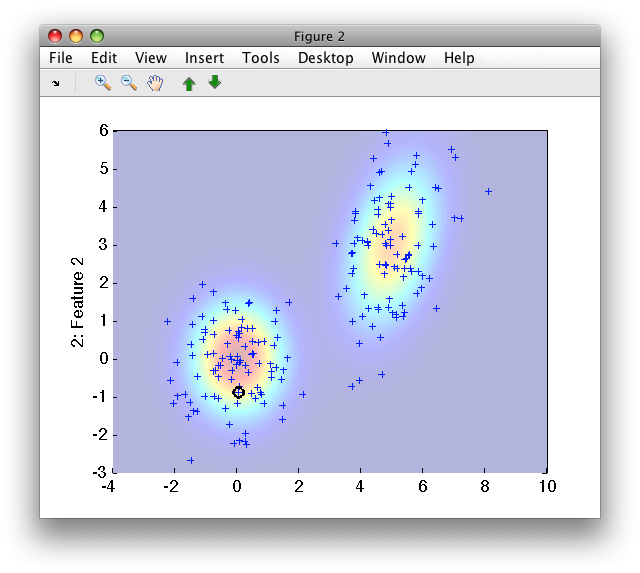
13.2.8. Gaussian mixture models ↩
Gaussian mixture is a density-estimation approach using a weighted sum of
multiple Gaussian components. The sdmixture implements a classifier with
a separate mixture model, estimated for each of the classes.
Advantage of a mixture classifier is its flexibility. Given enough training data, it may reliably model class distributions with arbitrary shapes or disjoint clusters (modes). Multi-modal data often originate in applications where our top-level classes reflect a composition of underlying states. For example, a medical diagnostic tool aims at detection of cancer. As the disease may affect different tissues, we observe our "cancer" class as a multi-modal composition of separate data tissue clusters. Mixture classifier can naturally describe such class distribution.
While parameters of a single Gaussian model may be estimated directly from data, mixture models require iterative optimization based on EM algorithm.
13.2.8.1. Automatic estimation of number of mixture components ↩
In perClass, the sdmixture command does not need any input arguments
apart of training data:
>> a
'Fruit set' 260 by 2 sddata, 3 classes: 'apple'(100) 'banana'(100) 'stone'(60)
>> p=sdmixture(a)
[class 'apple' init:.......... 4 clusters EM:done 4 comp] [class 'banana' init:....
...... 4 clusters EM:done 4 comp] [class 'stone' init:.......... 2 clusters EM:done 2 comp]
sequential pipeline 2x1 'Mixture of Gaussians+Decision'
1 Mixture of Gaussians 2x3 10 components, full cov.mat.
2 Decision 3x1 weighting, 3 classes
>> sdscatter(a,p)
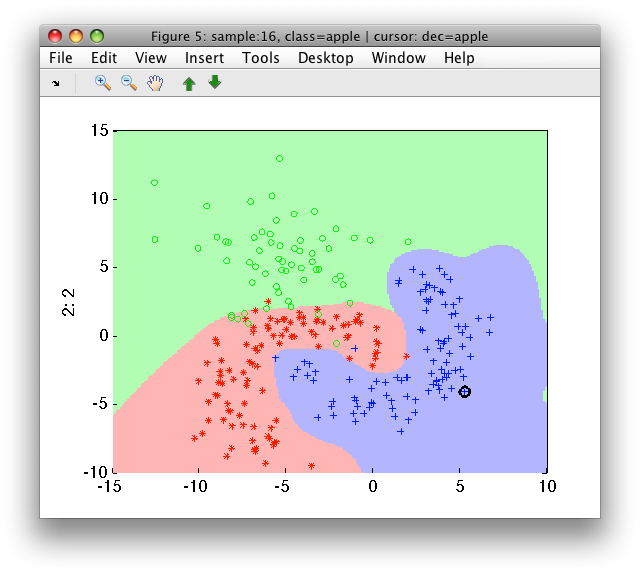
By default, sdmixture automatically identifies the number of components
for each class and then estimates each per-class mixture running 100
iterations of EM algorithm. Number of iterations can be changed with 'iter'
option.
The estimation of the number of components performs an internal random
split of the input data set. Therefore, we will receive potentially
different solution each run. Fixing the seed of the Matlab random number
generator makes the sdmixture training repeatable.
>> rand('state',1234); p=sdmixture(a) % example of setting random seed
Let us visualize the soft outputs of the mixture. We will use -p
shorthand to remove the decision step:
>> sdscatter(a,-p)
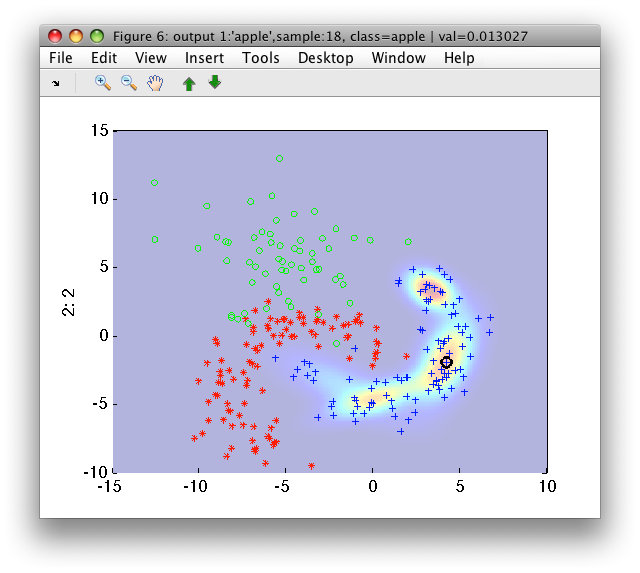
We can see the density of the mixture model for the first class ('apple') comprised of four components. Clicking the arrow buttons on the Figure toolbar, we can see the density estimates for the second and third class
The estimation of number of components is performed on a subset of the input data. By default 500 samples is used per class. This may be adjusted using 'maxsamples' option.
The algorithm uses a grid search considering 1:10 clusters by default. The grid may be adjusted with 'cluster grid' option.
13.2.8.2. Choosing number of mixture components manually ↩
We may fix the number of mixture components manually by providing it as the second argument:
>> p=sdmixture(a,5)
[class 'apple'EM:done 5 comp] [class 'banana'EM:done 5 comp] [class 'stone'EM:done 5 comp]
sequential pipeline 2x1 'Mixture of Gaussians+Decision'
1 Mixture of Gaussians 2x3 15 components, full cov.mat.
2 Decision 3x1 weighting, 3 classes
If a scalar value is given, the same number of components is used for all classes.
By providing a vector, we may fix different number of components per class. In our example, we know that the 'stone' class is unimodal:
>> p=sdmixture(a,[5 5 1])
[class 'apple'EM:done 5 comp] [class 'banana'EM:done 5 comp] [class 'stone'EM:done 1 comp]
sequential pipeline 2x1 'Mixture of Gaussians+Decision'
1 Mixture of Gaussians 2x3 11 components, full cov.mat.
2 Decision 3x1 weighting, 3 classes
13.2.8.3. Clustering data using a mixture model ↩
Mixture model which estimates the number of clusters automatically is a powerful data description tool. We may use it to cluster a data set i.e. to identify its distinct modes.
Let us consider a data set b with only one class. We create it by
removing class labels from fruit data set:
>> a
'Fruit set' 260 by 2 sddata, 3 classes: 'apple'(100) 'banana'(100) 'stone'(60)
>> b=sddata(+a)
260 by 2 sddata, class: 'unknown'
Let us train a mixture model twice. First, without any option:
>> rand('state',10); pc1=sdmixture(b)
[class 'unknown' init:.......... 8 clusters EM:done 8 comp]
sequential pipeline 2x1 'Mixture of Gaussians+Decision'
1 Mixture of Gaussians 2x1 8 components, full cov.mat.
2 Decision 1x1 threshold on 'unknown'
Second, with 'cluster' option:
>> rand('state',10); pc2=sdmixture(b,'cluster')
[class 'unknown' init:.......... 8 clusters EM:done 8 comp]
sequential pipeline 2x1 'Mixture of Gaussians+Decision'
1 Mixture of Gaussians 2x8 8 components, full cov.mat.
2 Decision 8x1 weighting, 8 classes
The rand commands make sure both estimated mixtures are identical.
The only difference between pipeline pc1 and pc2 is in labeling of the
mixture components.
The mixture pc1 models all available data in b by one mixture model
returning one soft output (see the output dimensionality of 1 in the
pipeline step 1). Let us visualize the decisions and soft outputs of pc1:
>> sdscatter(b,pc1)
ans =
1
>> sdscatter(b,-pc1)
ans =
2
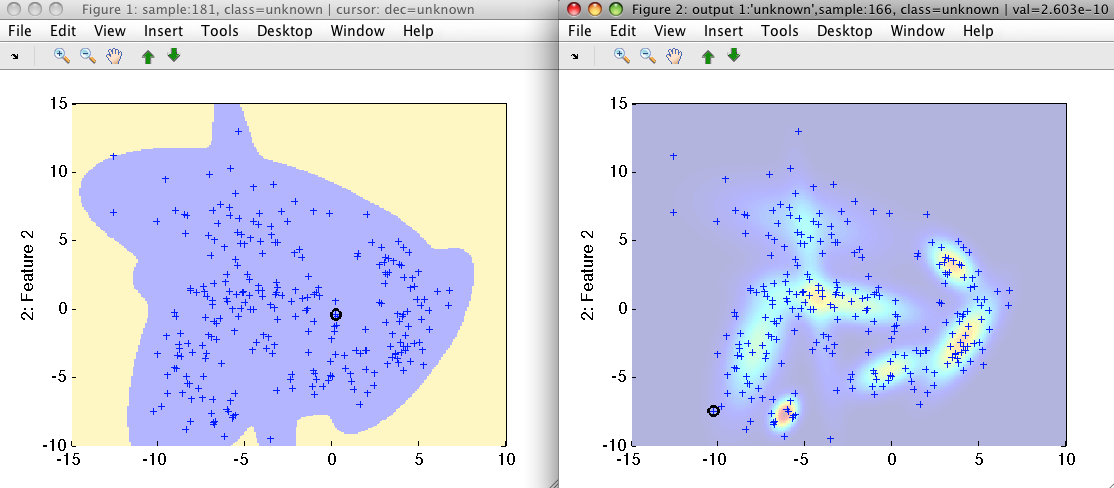
The pc2 pipeline, trained with 'cluster' option, splits the complete
mixture into individual components. Therefore, we can see 8 distinct soft
output. Default decisions will be 'Cluster 1' to 'Cluster 8':
>> sdscatter(b,pc2)
ans =
3
>> sdscatter(b,-pc2)
ans =
4
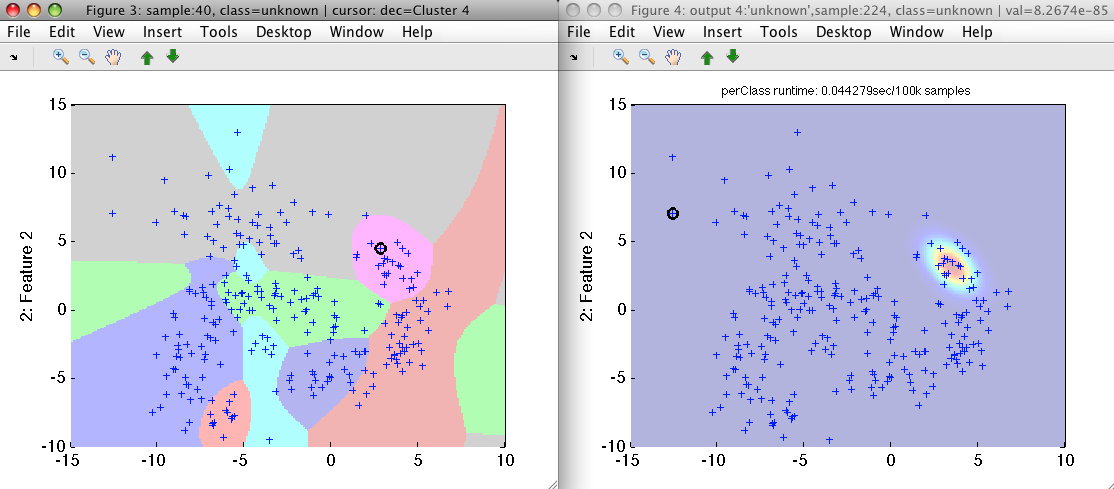
Note that we observe only one component at a time in the soft output Figure
4. Others are available via arrow buttons. The decisions of pc2 provide
cluster labels.
13.2.9. Regularization of Gaussian models ↩
Regularization is a technique of avoiding problem with parameter estimation caused by with small number of data samples in a problem with large dimensionality.
Normal-based classifier suffer, in such situations, from low quality estimates of class covariances. This results either in poor classifier performance or failure to train on a given data set.
Possible solution is to add a small constant to diagonal elements of the covariance matrices. In perClass, this can be achieved with the 'reg' option.
In the following example, we want to recognize handwritten digits represented directly by pixels in 16x16 raster. Quadratic classifier fails to discriminate the digits if trained on our limited training set of 60 samples per class. The full covariances are not reliably estimated.
>> [tr,ts]=randsubset(a,0.3)
'Digits' 600 by 256 sddata, 10 classes: [60 60 60 60 60 60 60 60 60 60]
'Digits' 1400 by 256 sddata, 10 classes: [140 140 140 140 140 140 140 140 140 140]
>> p=sdquadratic(tr)
sequential pipeline 256x1 'Quadratic discriminant'
1 Gaussian model 256x10 full cov.mat.
2 Normalization 10x10
3 Decision 10x1 weighting, 10 classes
>> sdtest(ts,p)
ans =
0.9000
Using the 'reg' option, sdquadratic performs automatic choice of the
regularization parameter. The test set error of the resulting classifier is
only 8.5%!
>> p=sdquadratic(tr,'reg')
..........
reg=0.100000 err=0.07
sequential pipeline 256x1 'Quadratic discriminant'
1 Gaussian model 256x10 full cov.mat.
2 Normalization 10x10
3 Decision 10x1 weighting, 10 classes
>> sdtest(ts,p)
ans =
0.0921
The 'reg' option splits internally the provided data into two parts and performs a grid search. One part is used for training the model, the other one for evaluating the error. The default splitting fraction is 20% for validation. It may be changed with the 'tsfrac' option.
Alternatively, the internal splitting is avoided by providing the test (validation) set with 'test' option:
>> rand('state',42); [tr2,val]=randsubset(tr,0.5)
>> p=sdquadratic(tr2,'reg','test',val);
The regularization parameter may be provided directly:
>> p=sdquadratic(tr,'reg',0.01)
