Classifiers, table of contents
This section describes Parzen density estimation and classification.
- 13.4.1. Introduction
- 13.4.2. Adjusting smoothing parameter manually
- 13.4.3. Vector smoothing
13.4.1. Introduction ↩
Parzen classifier estimates probability density for each class using a non-parametric approach based on stored training examples. When computing output for a new observation, the contribution of each training example is integrated. The contribution is modeled by a kernel function and is influenced by the smoothing parameter (kernel width).
By default, sdparzen trains a Parzen classifier with Laplace kernel
function which is less computationally demanding than frequently-adopted
Gaussian kernel. Smoothing parameter is optimized using EM algorithm
optimizing cross-validated log-likelihood.
>> load fruit
260 by 2 sddata, 3 classes: 'apple'(100) 'banana'(100) 'stone'(60)
>> p=sdparzen(a)
.....sequential pipeline 2x1 'Parzen model+Decision'
1 Parzen model 2x3 260 prototypes, h=0.8
2 Decision 3x1 weighting, 3 classes
>> sdscatter(a,p)
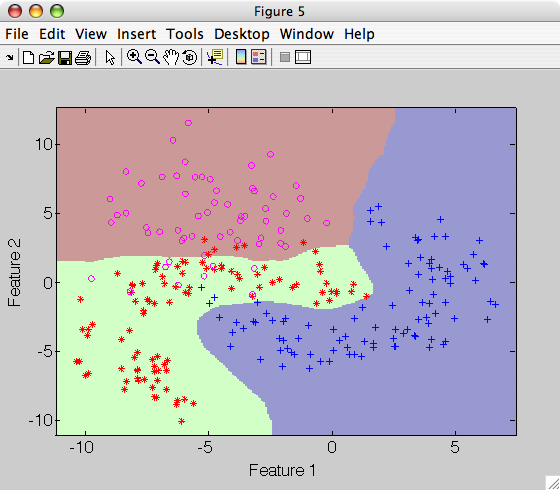
The default Parzen classifier uses scalar smoothing i.e. equal kernel width
for each feature. Smoothing parameter is accessible in h field of Parzen
pipeline:
>> p(1)'
Parzen model pipeline 2x3
1 Parzen model 2x3 260 prototypes, h=0.8
inlab: '1','2'
lab: 'apple','banana','stone'
output: probability density
h: smoothing parameter
13.4.2. Adjusting smoothing parameter manually ↩
Smoothing may be fixed manually. This is advantageous for quick experimentation because it skips the time-consuming EM algorithm.
Here we use very small kernel with:
>> p=sdparzen(a,'h',0.04)
>> sdscatter(a,p)
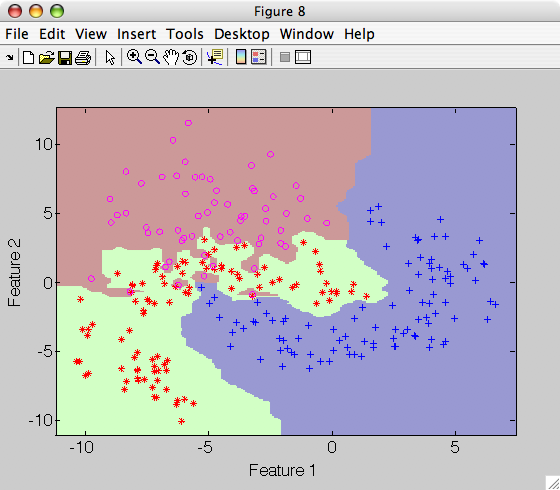
Note that the decision boundary becomes very complicated emphasizing very local changes of the class distributions.
13.4.3. Vector smoothing ↩
Smoothing parameter may be also estimated for each dimension, specifying
h as vector:
>> p=sdparzen(a,'h','vector')
.............sequential pipeline 2x1 'Parzen model+Decision'
1 Parzen model 2x3 260 prototypes, vector smoothing
2 Decision 3x1 weighting, 3 classes
>> p(1).h
ans =
0.6265 1.0200
Vector smoothing requires extra multiplication for each dimension of each
training sample. Alternative strategy is to scale the data to unit variance
with sdscale so that scalar smoothing is sufficient.
