Classifiers, table of contents
This section describes decision trees and random forest classifiers.
- 13.8.1. Decision tree classifier
- 13.8.1.1. Growing tree without pruning
- 13.8.1.2. Controlling the tree pruning
- 13.8.1.3. Using decision tree for feature selection
- 13.8.2. Random forest classifier
13.8.1. Decision tree classifier ↩
perClass sdtree classifier offers a fast and scalable decision tree
implementation. Decision tree is build by finding the best threshold on one
of the features that improves class separation. The process is applied
recursively until the stopping condition is met. If the tree is fully
built, each data sample ends in a separate terminal node. This solution,
however, does not yield good generalization in case of class overlap. As
with other classifiers, perClass strives to provide good solution by
default. Therefore, sdtree applies pruning strategy that stops tree
growth at the earlier stage. The pruning uses a separate validation set to
estimate tree generalization error.
To illustrate the basic use of the decision tree, lets consider the
fruit_large data set.
>> load fruit_large
>> a
'Fruit set' 2000 by 2 sddata, 3 classes: 'apple'(667) 'banana'(667) 'stone'(666)
We split the data to training and test subsets. The test subset will be used later to estimate tree performance.
>> [tr,ts]=randsubset(a,0.5)
'Fruit set' 999 by 2 sddata, 3 classes: 'apple'(333) 'banana'(333) 'stone'(333)
'Fruit set' 1001 by 2 sddata, 3 classes: 'apple'(334) 'banana'(334) 'stone'(333)
We train the tree on the training subset tr. By default, the data set
passed to sdtree is split internally into two subsets. The first part is
used to grow the tree and the second part to limit its growth by
identifying the sufficient number of thresholds. This process happens
inside sdtree. We will see later, how to take closer control over the
pruning process.
>> p=sdtree(tr)
sequential pipeline 2x1 'Decision tree'
1 Decision tree 2x3 14 thresholds on 2 features
2 Decision 3x1 weighting, 3 classes
Let's visualize the decisions at the default operating point on the test set:
>> sdscatter(ts,p)
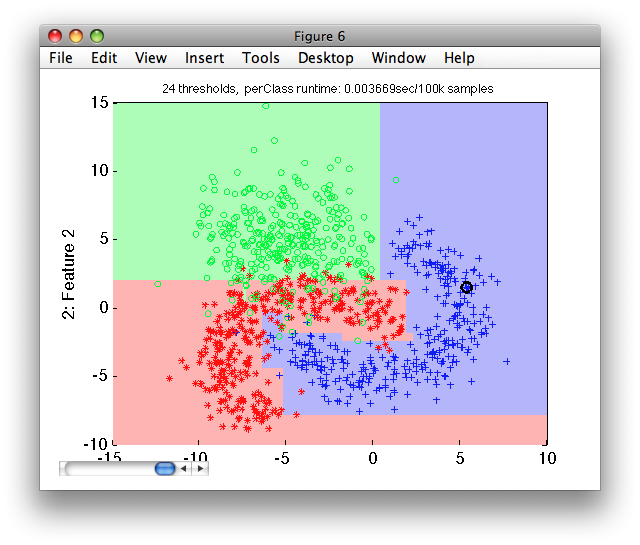
We now estimate the mean test error using sdtest on the test subset:
>> sdtest(ts,p)
ans =
0.0779
13.8.1.1. Growing tree without pruning ↩
We may suppress the pruning process and grow the full tree, using the 'full' option.
>> p2=sdtree(tr,'full');
>> p2'
sequential pipeline 2x1 'Decision tree'
1 Decision tree 2x3 97 thresholds on 2 features
inlab: 'Feature 1','Feature 2'
lab: 'apple','banana','stone'
output: posterior
thresholds: number of thresholds
feat: features used
2 Decision 3x1 weighting, 3 classes
inlab: 'apple','banana','stone'
output: decision ('apple','banana','stone')
Inspecting the pipeline steps, we can see that the number of thresholds and the features used are available for direct query. Note how the number of thresholds is much higher than when pruning the tree.
>>[p(1).thresholds p2(1).thresholds]
ans =
14 97
We may compare the fully grown tree with the tree built using the default pruning strategy:
>> sdscatter(ts,p2)
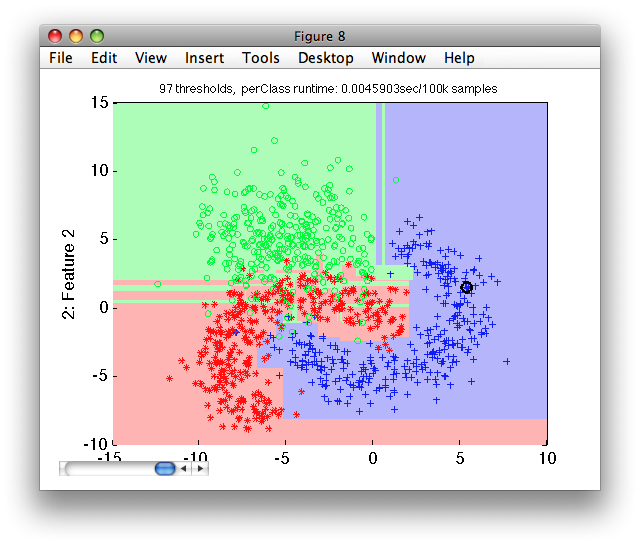
Notice, the over-fitting appearing in the area of overlap. The fully-grown tree solves perfectly separates the training set but fails to do so on the independent test set.
13.8.1.2. Controlling the tree pruning ↩
We may control the sdtree pruning algorithm in several ways. Firstly, we
may specify the fraction of the input data set used for training the tree
using the 'trfrac' option. By default, 80% of input data is used to grow
the tree and 20% to stop the growth process.
>> p=sdtree(tr,'trfrac',0.5)
sequential pipeline 2x1 'Decision tree'
1 Decision tree 2x3 11 thresholds on 2 features
2 Decision 3x1 weighting, 3 classes
Secondly, we may remove the random splitting step by supplying the separate validation data used for pruning.
>> [val,tr2]=randsubset(tr,100)
'Fruit set' 300 by 2 sddata, 3 classes: 'apple'(100) 'banana'(100) 'stone'(100)
'Fruit set' 699 by 2 sddata, 3 classes: 'apple'(233) 'banana'(233) 'stone'(233)
We train on the tr2 part and pass the validation set val separately
using the 'test' option:
>> p=sdtree(tr2,'test',val)
sequential pipeline 2x1 'Decision tree'
1 Decision tree 2x3 11 thresholds on 2 features
2 Decision 3x1 weighting, 3 classes
This removes the random element from the sdtree training. Repeating the
training, we obtain identical tree.
>> p2=sdtree(tr2,'test',val)
sequential pipeline 2x1 'Decision tree'
1 Decision tree 2x3 11 thresholds on 2 features
2 Decision 3x1 weighting, 3 classes
>> [sdtest(ts,p) sdtest(ts,p2)]
ans =
0.0829 0.0829
Thirdly, the number of tree levels (thresholds) may be also specified using the 'levels' option during the tree training:
>> p=sdtree(tr,'levels',10);
>> p(1).thresholds
ans =
10
We may also inspect the pruning process in detail. The second output
parameter, returned by sdtree, is a structure containing the full tree
and the error criterion estimated from the validation set at each tree
threshold.
>> [p,res]=sdtree(tr2,'test',val)
sequential pipeline 2x1 'Decision tree'
1 Decision tree 2x3 11 thresholds on 2 features
2 Decision 3x1 weighting, 3 classes
res =
full_tree: [2x3 sdppl]
err: [42x1 int32]
>> figure; plot(res.err)
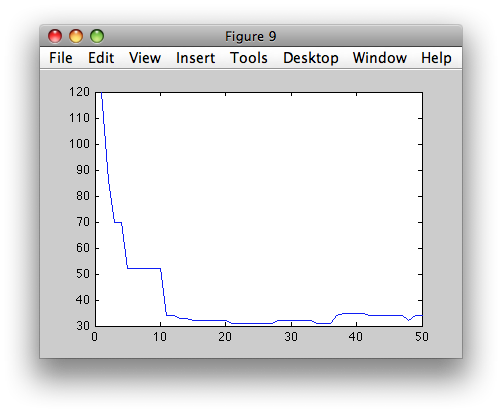
The res.full_tree field contains the fully grown tree. To be more
precise, it contains the tree grown as much as possible depending on the
'maxsamples' option. By default, at least 10 samples much be present in a
node to continue growing process.
To prune the tree manually, we may select a specific number of thresholds
by passing tree pipeline to the sdtree function. Here, we select 10
thresholds:
>> p3=sdtree(p,10)
sequential pipeline 2x1 'Decision tree+Decision'
1 Decision tree 2x3 10 thresholds on 2 features
2 Decision 3x1 weighting, 3 classes
13.8.1.3. Using decision tree for feature selection ↩
Decision trees optimize class separability considering all features. Therefore, a trained tree allows us to identify the features that provided most separability in our problem. In other words, we may use it for feature selection.
In this example, we train a tree classifier on medical data set:
>> a
'medical D/ND' 5762 by 10 sddata, 2 classes: 'disease'(1495) 'no-disease'(4267)
>> p=sdtree(a)
sequential pipeline 10x1 'Decision tree'
1 Decision tree 10x2 41 thresholds on 8 features
2 Decision 2x1 weighting, 2 classes
You may see in the pipeline comment, that only 8 features form 10 is used in this tree. Removing the unused features allows us to limit the amount of computation we need in our final system.
By passing the tree to sdfeatsel function, we obtain a pipeline
describing the same classifier but only on these 8 features:
>> pf=sdfeatsel(p)
sequential pipeline 10x1 'Feature subset+Decision tree+Decision'
1 Feature subset 10x8
2 Decision tree 8x2 41 thresholds on 8 features
3 Decision 2x1 weighting, 2 classes
The first step in the pipeline pf is the feature selection. We may list
the features that are useful:
>> +pf(1).lab
ans =
StdDev
Skew
Kurtosis
Energy Band 1
Energy Band 2
Moment 20
Moment 02
Fractal 4
Typically, we would use this first step in the pipeline pf to create a new
data set with only relevant features:
>> b=a*pf(1)
'medical D/ND' 5762 by 8 sddata, 2 classes: 'disease'(1495) 'no-disease'(4267)
The classifier pf(2:end) is running on this data set:
>> dec=b*pf(2:end)
sdlab with 5762 entries, 2 groups: 'disease'(1459) 'no-disease'(4303)
13.8.2. Random forest classifier ↩
Random forest classifier sdrandforest combines a large number of
specifically-built decision trees. Each tree is built by considering only
randomly selected subset of features at each tree node. The combination of
their outputs is based on the sum rule.
By default, sdrandforest builds 20 trees and considers a subset of 20% of
features at each node. Let us train a random forest classifier on the
medical data set. We use data of the first five patients as a test set and
remaining data as a training set:
>> load medical
>> a=a(:,:,1:2)
'medical D/ND' 5762 by 11 sddata, 2 classes: 'disease'(1495) 'no-disease'(4267)
>> [ts,tr]=subset(a,'patient',1:5)
'medical D/ND' 1887 by 11 sddata, 2 classes: 'disease'(597) 'no-disease'(1290)
'medical D/ND' 3875 by 11 sddata, 2 classes: 'disease'(898) 'no-disease'(2977)
>> p=sdrandforest(tr)
..........
sequential pipeline 11x1 'Random forest+Decision'
1 Random forest 11x2 20 trees
2 Decision 2x1 weighting, 2 classes
>> sdtest(ts,p)
ans =
0.2730
sdrandforest offers several user-adjustable parameters. For example, we
may alter the number of trees and the number of randomly-selected
dimensions.
The number of trees may be provided directly as the second parameter:
>> p=sdrandforest(tr,200)
..........
sequential pipeline 11x1 'Random forest+Decision'
1 Random forest 11x2 200 trees
2 Decision 2x1 weighting, 2 classes
>> sdtest(ts,p)
ans =
0.2848
To adjust the number of dimensions that are randomly selected at each tree node during training, use the 'dim' option. Here we combine it with the request for the 200 trees:
>> p=sdrandforest(tr,'dim',0.5,'count',200)
..........
sequential pipeline 11x1 'Random forest+Decision'
1 Random forest 11x2 200 trees
2 Decision 2x1 weighting, 2 classes
>> sdtest(ts,p)
ans =
0.2798
