Classifiers, table of contents
This section describes classifiers based on k-th nearest neighbor rule.
- 13.3.1. k-NN classifier
- 13.3.1.1. Changing neighborhood size k
- 13.3.1.2. Data scaling
- 13.3.1.3. Different k-NN formulations
- 13.3.2. Prototype selection for k-NN
- 13.3.3. k-means as a classifier
- 13.3.3.1. Adjusting number of centers in the k-means classifier
- 13.3.3.2. Changing k in the final k-NN classifier
- 13.3.3.3. Prototype pruning
13.3.1. k-NN classifier ↩
k-NN (k-th nearest neighbor) is a non-parametric classifier measuring distance between a new sample and stored "prototype" examples. Instead of building a model from training examples it uses them directly as evidence for computing its output on new observations.
By default sdknn implements the first nearest neighbor rule (k=1) using all
provided training examples as prototypes:
>> load fruit
>> a
'Fruit set' 260 by 2 sddata, 3 classes: 'apple'(100) 'banana'(100) 'stone'(60)
>> p=sdknn(a)
sequential pipeline 2x1 '1-NN+Decision'
1 1-NN 2x3 260 prototypes
2 Decision 3x1 weighting, 3 classes
>> sdscatter(a,p)
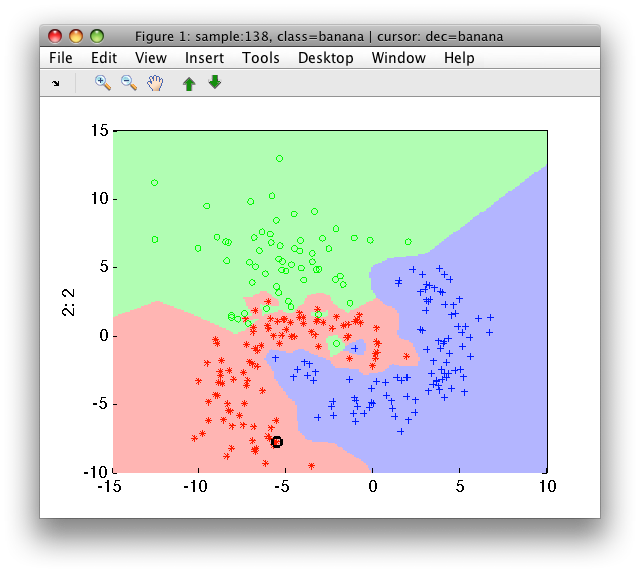
Using larger neighborhoods (k>1), the nearest neighbor classifier becomes
more robust against noise in the areas of overlap.
13.3.1.1. Changing neighborhood size k ↩
We may specify the neighborhood size k as second parameter:
>> p3=sdknn(a,3)
sequential pipeline 2x1 '3-NN+Decision'
1 3-NN 2x3 260 prototypes
2 Decision 3x1 weighting, 3 classes
>> sdscatter(a,p3)
An alternative is to directly change the k field in the pipeline step:
>> p3'
sequential pipeline 2x1 '3-NN+Decision'
1 3-NN 2x3 260 prototypes
inlab: '1','2'
lab: 'apple','banana','stone'
output: distance
k: number of neighbors
2 Decision 3x1 weighting, 3 classes
inlab: 'apple','banana','stone'
output: decision ('apple','banana','stone')
>> p3(1).k=10
sequential pipeline 2x1 '10-NN+Decision'
1 10-NN 2x3 260 prototypes
2 Decision 3x1 weighting, 3 classes
13.3.1.2. Data scaling ↩
The k-NN classifier is based on Euclidean distance. This means that, if some of the features have significantly different scale than others, they dominate in the classification process. Therefore, we may wish to scale the data before using k-NN classifier.
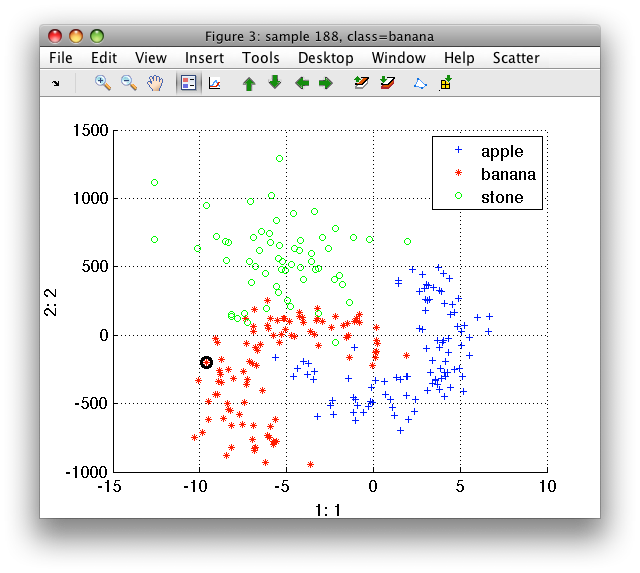
Lets make a fruit data set with second feature scaled by 100:
>> b=setdata(a, [+a(:,1) +a(:,2)*100])
'Fruit set' 260 by 2 sddata, 3 classes: 'apple'(100) 'banana'(100) 'stone'(60)
>> sdscatter(b)
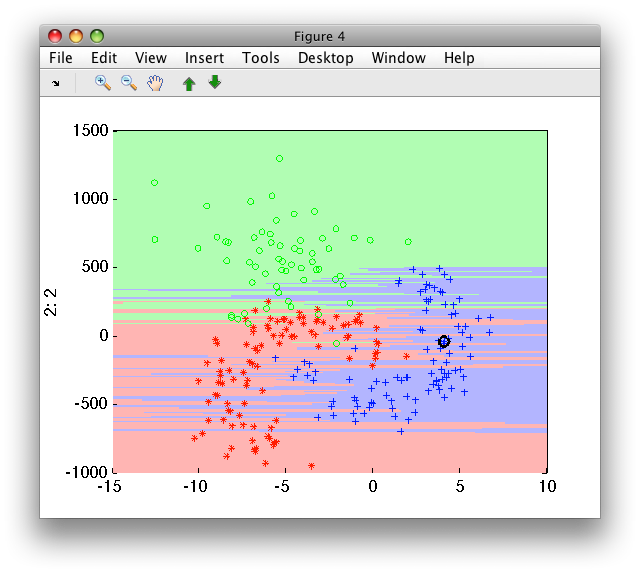
If we train k-NN on this data, the classifier decisions will be strongly affected by incomparable scales of the two features:
>> pk1=sdknn(b)
sequential pipeline 2x1 '1-NN+Decision'
1 1-NN 2x3 260 prototypes
2 Decision 3x1 weighting, 3 classes
>> sdscatter(b,pk1)
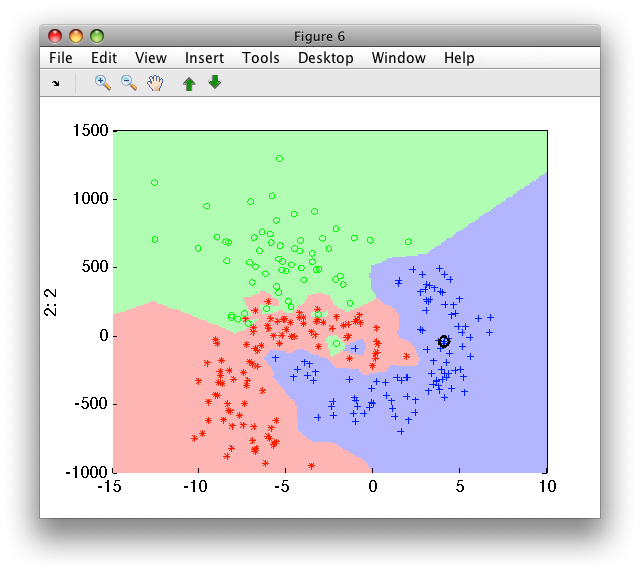
Training the k-NN on the scaled version makes both feature equally
important. The sdscale by default yields zero mean and unit variance:
>> ps=sdscale(b)
Scaling pipeline 2x2 standardization
>> pk2=sdknn(b*ps)
sequential pipeline 2x1 '1-NN+Decision'
1 1-NN 2x3 260 prototypes
2 Decision 3x1 weighting, 3 classes
>> sdscatter(b,ps*pk2)
Training of the k-NN on scaled data may be written on one line by constructing an untrained pipeline:
>> pu=sdscale*sdknn([],3)
untrained pipeline 2 steps: sdscale+sdknn
>> ptr=b*pu
sequential pipeline 2x1 'Scaling+3-NN+Decision'
1 Scaling 2x2 standardization
2 3-NN 2x3 260 prototypes
3 Decision 3x1 weighting, 3 classes
13.3.1.3. Different k-NN formulations ↩
By default, sdknn measures, for each class, a distance to the k-th
nearest prototype. An alternative formulation finds k nearest prototypes
irrespective of their classes and returns a fraction of each class among
these k prototypes. The nearest neighbor computing class fractions is
created by setting the method to classfrac:
>> p2=sdknn(a,10,'classfrac')
sequential pipeline 2x1 '10-NN discriminant+Decision'
1 10-NN discriminant 2x3 260 prototypes
2 Decision 3x1 weighting, 3 classes
Although both methods may look similar at the first sight, different way of computing the per-class output has important practical consequences. The distance method returns distance while the classfrac the similarity (fraction):
>> p1(1).output
ans =
distance
>> p2(1).output
ans =
posterior
Because classfrac is computing class fractions between k neighbors, it
provides a discriminant applicable to two or more classes. This means
that it splits the feature space into open sub-spaces. It separates classes
but cannot be used to detect one class protecting it from all directions.
The distance method, on the other hand, computes distance to k-th nearest
neighbor for each class separately:
>> p=sdknn( a(:,:,'banana') ,10,'classfrac')
{??? Error using ==> sdknn at 209
k-NN with classfrac option cannot be used for one-class problems.
}
>> p=sdknn( a(:,:,'banana') ,10)
sequential pipeline 2x1 '10-NN+Decision'
1 10-NN 2x1 100 prototypes
2 Decision 1x1 threshold on 'banana'
>> sdscatter(a,p)
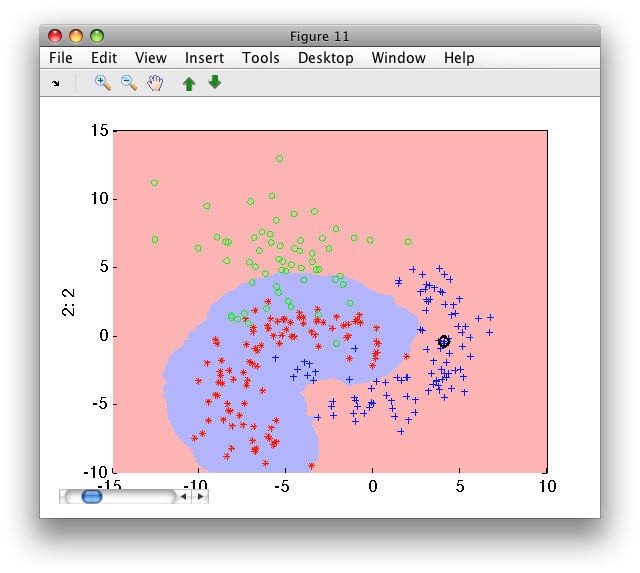
13.3.2. Prototype selection for k-NN ↩
By default sdknn uses all training examples as prototypes. It may,
however, also select a smaller subset. Two prototype selection strategies
are used:
- random - selecting a specified number of prototypes per class
- kcentres - using
sdkcentresclustering algorithm to systematically select prototypes.
In this example, we randomly select 40 prototypes per class:
>> p=sdknn(a,'k',5,'proto',40)
sequential pipeline 2x1 '5-NN+Decision'
1 5-NN 2x3 120 prototypes
2 Decision 3x1 weighting, 3 classes
Number of prototypes may be adjusted per class. Simpler classes may be described by smaller prototype sets. This increases execution speed of the k-NN classifier:
>> p=sdknn(a,'k',5,'proto',[40 40 10])
sequential pipeline 2x1 '5-NN+Decision'
1 5-NN 2x3 90 prototypes
2 Decision 3x1 weighting, 3 classes
To select prototypes using kcentres, use protosel option:
>> p=sdknn(a,'k',5,'proto',40,'protosel','kcentres')
[class 'apple' 40 comp]
[class 'banana' 40 comp]
[class 'stone' 40 comp]
sequential pipeline 2x1 '5-NN+Decision'
1 5-NN 2x3 120 prototypes
2 Decision 3x1 weighting, 3 classes
k-centres algorithm computes dissimilarity matrix. For large data sets,
sdknn uses only randomly drawn 2000 examples to compute k-centres.
13.3.3. k-means as a classifier ↩
k-means is a clustering approach where k cluster centers are derived by an iterative procedure minimizing mean (squared) distance of observation in each cluster.
perClass sdkmeans can be used for cluster analysis but is, by default,
also a classifier. It performs the cluster optimization for each of the
classes and outputs a 1-NN based-classifier. The prototypes of this
classifier are the cluster centers.
The advantage of using k-means as a classifier is the systematic reduction of number of prototypes compared to a k-NN classifier that employes all training examples. This yields classifiers of comparable performance but significantly faster in execution.
>> load fruit_large
>> a
'Fruit set' 2000 by 2 sddata, 3 classes: 'apple'(667) 'banana'(667) 'stone'(666)
>> p=sdkmeans(a,50)
[class 'apple' 13 iters. 9 clusters pruned, 41 clusters]
[class 'banana' 19 iters. 14 clusters pruned, 36 clusters]
[class 'stone' 18 iters. 10 clusters pruned, 40 clusters]
sequential pipeline 2x1 '1-NN (k-means prototypes)+Decision'
1 1-NN (k-means prototypes) 2x3 117 prototypes
2 Decision 3x1 weighting, 3 classes
>> sdscatter(randsubset(a,200),p)
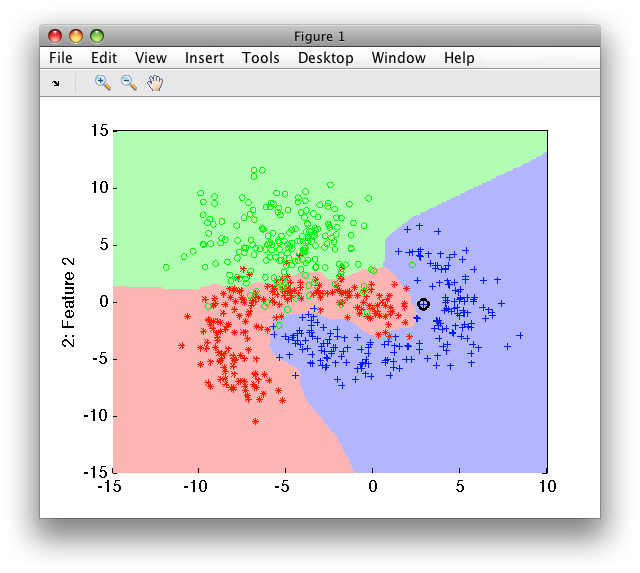
By default, 200 iterations is performed unless a stable solution is
reached. In the following example, we limit the number of iterations to
15. As we set identical random seed before running sdkmeans, we work
exactly on the same data with identical initialization as above. Due to
the iteration limit, we receive warning for the second and third classes:
>> rand('state',1); p=sdkmeans(a,50,'iter',15)
[class 'apple' 13 iters. 9 clusters pruned, 41 clusters] [class 'banana' 15 iters. {Warning: sdkmeans stopped without converging after the limit of 15 iterations.
Increase the max numbef of iterations using 'iter' option.}
14 clusters pruned, 36 clusters] [class 'stone' 15 iters. {Warning: sdkmeans stopped without converging after the limit of 15 iterations.
Increase the max numbef of iterations using 'iter' option.}
10 clusters pruned, 40 clusters]
sequential pipeline 2x1 '1-NN (k-means prototypes)+Decision'
1 1-NN (k-means prototypes) 2x3 117 prototypes
2 Decision 3x1 weighting, 3 classes
13.3.3.1. Adjusting number of centers in the k-means classifier ↩
Different k may be specified for each class. This is useful if we know that some classes are unimodal (like 'stone' on our example):
>> p=sdkmeans(a,[30 30 1])
[class 'apple' 6 clusters pruned, 24 clusters] [class 'banana' 10 clusters pruned, 20 clusters] [class 'stone' 1 clusters]
sequential pipeline 2x1 '1-NN (k-means prototypes)+Decision'
1 1-NN (k-means prototypes) 2x3 45 prototypes
2 Decision 3x1 weighting, 3 classes
13.3.3.2. Changing k in the final k-NN classifier ↩
By default, sdkmeans returns a 1-NN classifier. This may be changed by
the 'k final' option:
>> p=sdkmeans(a,50,'k final',2)
[class 'apple' 10 clusters pruned, 40 clusters] [class 'banana' 14 clusters pruned, 36 clusters] [class 'stone' 8 clusters pruned, 42 clusters]
sequential pipeline 2x1 '2-NN (k-means proto)+Decision'
1 2-NN (k-means proto) 2x3 118 prototypes
2 Decision 3x1 weighting, 3 classes
An alternative is to change the k field in the resulting pipeline:
>> p(1).k=10
sequential pipeline 2x1 '10-NN+Decision'
1 10-NN 2x3 118 prototypes
2 Decision 3x1 weighting, 3 classes
13.3.3.3. Prototype pruning ↩
By default, sdkmeans prunes prototypes in order to minimize
misclassification in the areas of overlap. Pruning may be disabled with 'no
pruning' option.
>> p=sdkmeans(a,50,'no pruning')
[class 'apple' 50 clusters] [class 'banana' 50 clusters] [class 'stone' 50 clusters]
sequential pipeline 2x1 '1-NN (k-means prototypes)+Decision'
1 1-NN (k-means prototypes) 2x3 150 prototypes
2 Decision 3x1 weighting, 3 classes
