Problem: How to combine image features extracted at different scales?
Solution: Use a simple syntax to work with a subset of pixels represented at all scales.
30.1. Introduction ↩
When classifying images, it is useful to extract features at different scales. While small scales describe object details, larger ones capture bigger structures.
Typically, we do not know which specific scale is optimal for our problem. Therefore, we need to compute features at multiple scales and combine them into one data set. With feature selection or extraction methods, we may derive a scale subset useful for our problem.
In this example, we demonstrate how to combine multi-scale features into one data set.
30.2. Computing image features at multiple scales ↩
Local image features, extracted from a square sliding window may be
computed with perClass sdextract command.
Let us create an image data set loading the dice image from data
sub-directory of perClass distribution:
>> a=sdimage('dice01.jpg','sddata')
101376 by 3 sddata, class: 'unknown'
The dice01.jpg image is loaded with Matlab imread command and reshaped
into sddata object a. Each pixel becomes one data sample and is
represented by the three features (R, G, and B).
We set the feature labels so that we may use the channel names:
>> a.featlab=sdlab('R','G','B')
101376 by 3 sddata, class: 'unknown'
To visualize our image, use sdimage command:
>> sdimage(a)
ans =
1
30.3. Constructing gray channel ↩
As we wish to work with a single gray-level channel, we create a new data
set gray fusing the information in the R,G, and B channels, respectively.
One way is to use setdata method to provide a new data matrix,
weighting the R, G and B channels:
>> gray=setdata( a, 0.29*+a(:,'R') + 0.587*+a(:,'G') + 0.11*+a(:,'B') )
101376 by 1 sddata, class: 'unknown'
Note the +a(:,'R') syntax that returns raw data matrix for the specified
channel.
An alternative is to create an affine projection pipeline and apply it to
the data set a:
>> p=sdaffine([0.29 0.587 0.11]',0)
Affine projection pipeline 3x1
>> gray=a*p
101376 by 1 sddata, class: 'unknown'
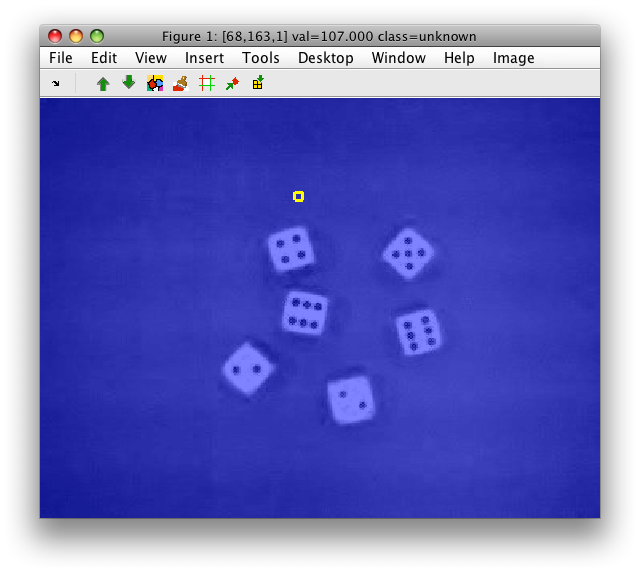
30.4. Smoothing image with Gaussian kernel ↩
We will now smooth the image with Gaussian kernel to access image information at larger scale:
>> g=sdextract(gray,'region','gauss','sigma',2,'block',15)
block: 15, sigma: 2.0 yields kernel coverage 100.0%
92612 by 1 sddata, class: 'unknown'
Note, that we specify both the filter sigma and the block size and receive a message computing the coverage of our Gaussian in the selected sliding window (for details see documentation)
It is important to note, that perClass computes region features only in
sliding windows that fully cover existing image data. Therefore, the number
of samples in the extracted data set g is smaller that in the original
gray image.
When we visualize the image g, we will notice the border along the image
edges where the Gaussian kernel could not be computed.
>> sdimage(g)
ans =
2
30.5. Concatenating images at different scales ↩
The original image data set gray and Gaussian-smoothed image g contain
different number of samples:
>> gray
101376 by 1 sddata, class: 'unknown'
>> g
92612 by 1 sddata, class: 'unknown'
Therefore, we cannot simply horizontally concatenate them:
>> data=[gray g]
??? Error using ==> sddata.horzcat at 56
Sample sizes do not match
What we can do, however, is to concatenate subset of pixels represented by both data sets.
perClass 4.3 introduces a simple syntax to find a subset of pixels represented in two image sets:
>> sub=gray(g)
92612 by 1 sddata, class: 'unknown'
The data set sub is created as a subset of data set gray with pixels
present both in gray and g.
We can now simply write:
>> data=[sub g]
92612 by 2 sddata, class: 'unknown'
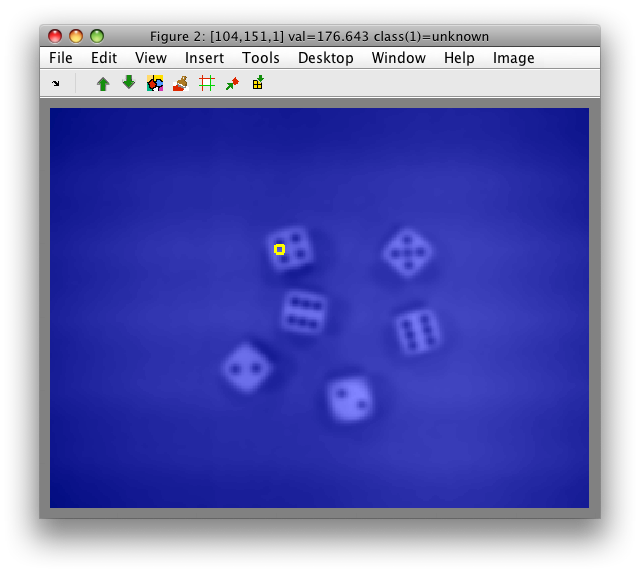
The resulting data set data is an image with two bands. You may visualize it with:
>> sdimage(data)
Flip between the two bands with up/down cursor keys or via toolbar buttons.
To show both bands in one image, we open two separate figures:
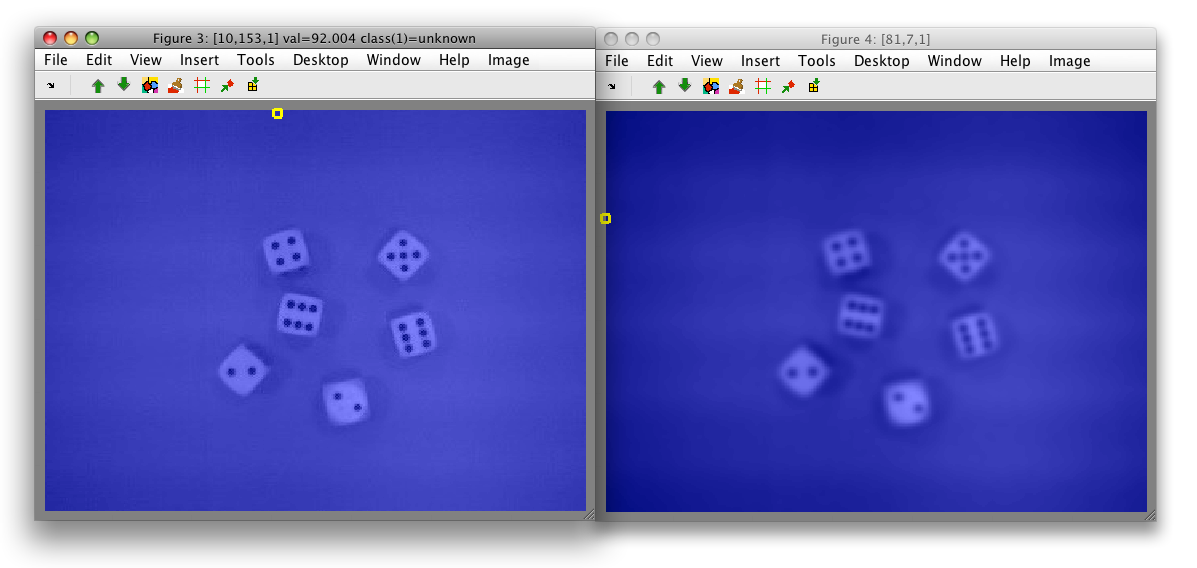
Note, that both figures show the identical gray border without data. This
is because the new data set data only contains pixels represented in both
original images.
30.6. Fusing images at multiple scales ↩
If we wish to concatenate images at multiple scales, we simply use preserve only pixels available in the band with largest scale.
For example, say we compute another Gaussian smoothed image, now with sigma of 5.0 in 31x31 windows:
>> g2=sdextract(gray,'region','gauss','sigma',5,'block',31)
block: 31, sigma: 5.0 yields kernel coverage 99.6%
83076 by 1 sddata, class: 'unknown'
The concatenation would then be:
>> data=[gray(g2) g(g2) g2]
83076 by 3 sddata, class: 'unknown'
Both, the gray original image and smaller-scale g image need to be
sub-sampled preserving only the pixels available in g2.
30.7. Passing labels from multi-scale to the original image ↩
When working with data sets containing image features at multiple scales, we often face the task of passing the image labels back to the original image.
For example, we may use the concatenated data set data from the example
above to define dice labels by clustering.
- Open the image view:
>> close all
>> sdimage(data)
- Select 'Cluster with k-means' from Image menu (or press 'k').
- Fill the desired number of clusters, say 3 and press OK.
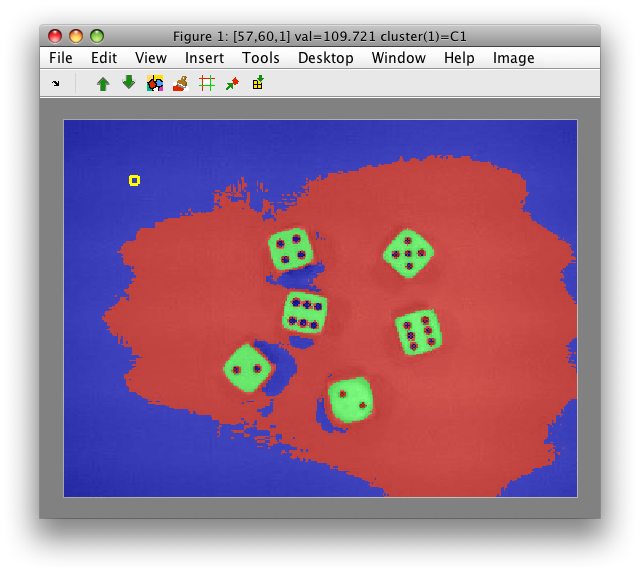
- The derived cluster labels are
C1,C2andC3. We will assign them meaningful names by the rename command. Press 'r' when hovering with the mouse over specific region and use class such as names 'dice' and 'back'. When you provide the same class name 'back' to the two background regions, they will be merged.
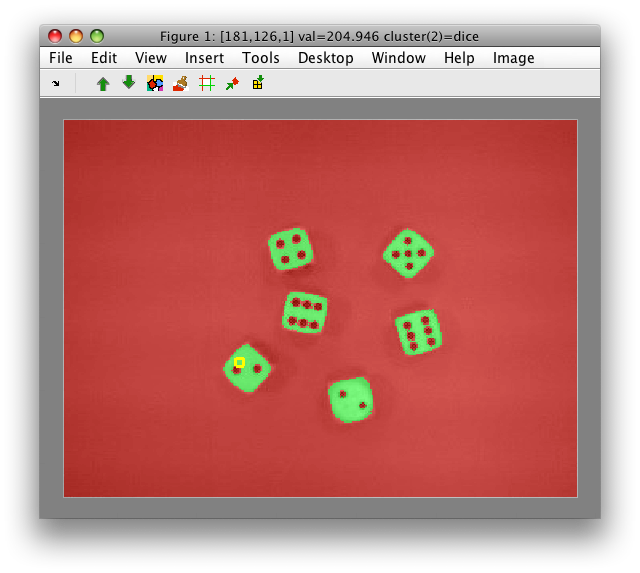
- Save the labels into Matlab workspace with 'Create current labels in workspace' command. use
Las the name of new label object
>> Creating label set L in the workspace.
sdlab with 83076 entries, 2 groups: 'back'(79736) 'dice'(3340)
The original image set a contains more pixels than the merged set data:
>> data
83076 by 3 sddata, class: 'unknown'
>> a
101376 by 3 sddata, class: 'unknown'
We may assign the labels L to a with:
>> a(data).lab=L
101376 by 3 sddata, 3 classes: 'unknown'(18300) 'back'(79736) 'dice'(3340)
>> sdimage(a)
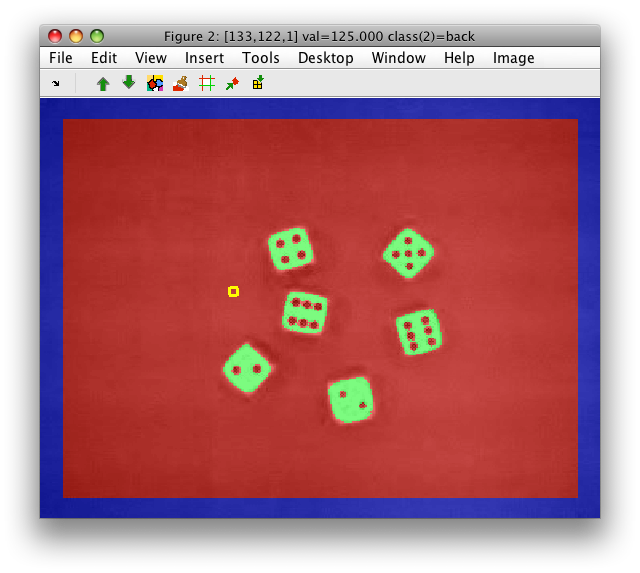
We have now passed the labels to the original image. Note, that apart of 'dice' and 'back' classes, there is also the 'unknown' class at the image border.
30.8. Passing labels from the original image to the multi-scale subset ↩
The second use-case is to pass labels from the original image to the
multi-scale subset. Say, we have hand-painted labels in the original RGB
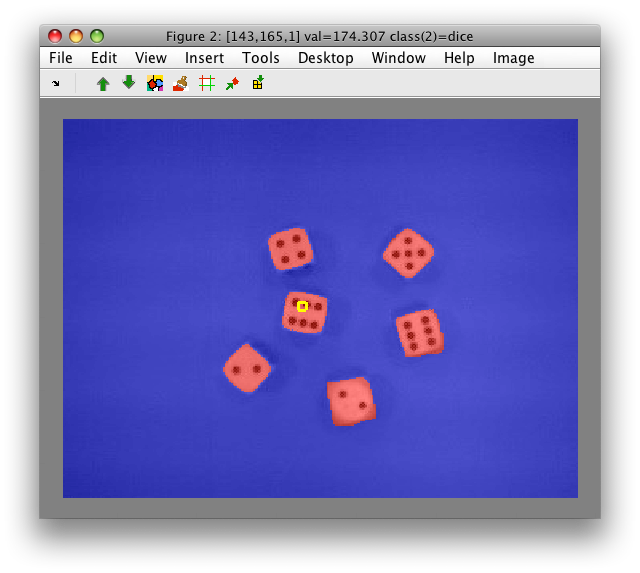
image a:
>> Creating data set a in the workspace.
101376 by 3 sddata, 2 classes: 'unknown'(97199) 'dice'(4177)
>> sdimage(a)
ans =
1
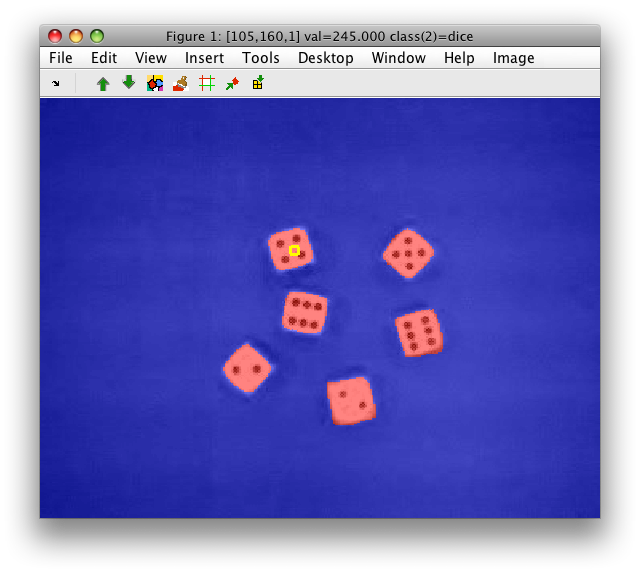
Our data set data contains gray-level band and two Gaussian-convolved
images at different scales:
>> data
83076 by 3 sddata, class: 'unknown'
>> data.featlab'
1 Output
2 Output,Gauss(sigma=2.00)
3 Output,Gauss(sigma=5.00)
To pass the labels a.lab to data, we first create a subset of the
original image with pixels that are represented in our multi-scale set
data:
>> sub=a(data)
83076 by 3 sddata, 2 classes: 'unknown'(78899) 'dice'(4177)
Now, we can assign the labels of sub into data:
>> data.lab=sub.lab
83076 by 3 sddata, 2 classes: 'unknown'(78899) 'dice'(4177)
>> sdimage(data)
ans =
2